Microbiomes
we offer microbiome analyses from various samples such as stool, soil, skin or buccal swabs. Besides 16S RNA gene sequencing (16S RNA-Seq, 16 s rRNA Sequencing) we offer ITS sequencing and metagenomics by shotgun sequencing of the DNA mixture of the sample.
Metagenomics
Metagenomics by shotgun sequencing is the analysis of DNA fragments derived from all organisms present in a sample is the most comprehensive method to analyse for example the human microbiome.
To obtain a representative spectrum of the available DNA, the samples must be carefully processed to obtain a representative portion of each genome. We apply whole-metagenome shotgun (WMS) sequencing or alternatively NGS of 16s-RNA coding regions.
The obtained sequence reads are annotated with high-performance computer clusters to all suitable sequences in the databases to identify the species present in the sample. Reads that cannot be annotated to database-entries can be de novo assembled to identify new genomes.
We provide a complete list of all identified organisms and their relative frequencies in the sample.
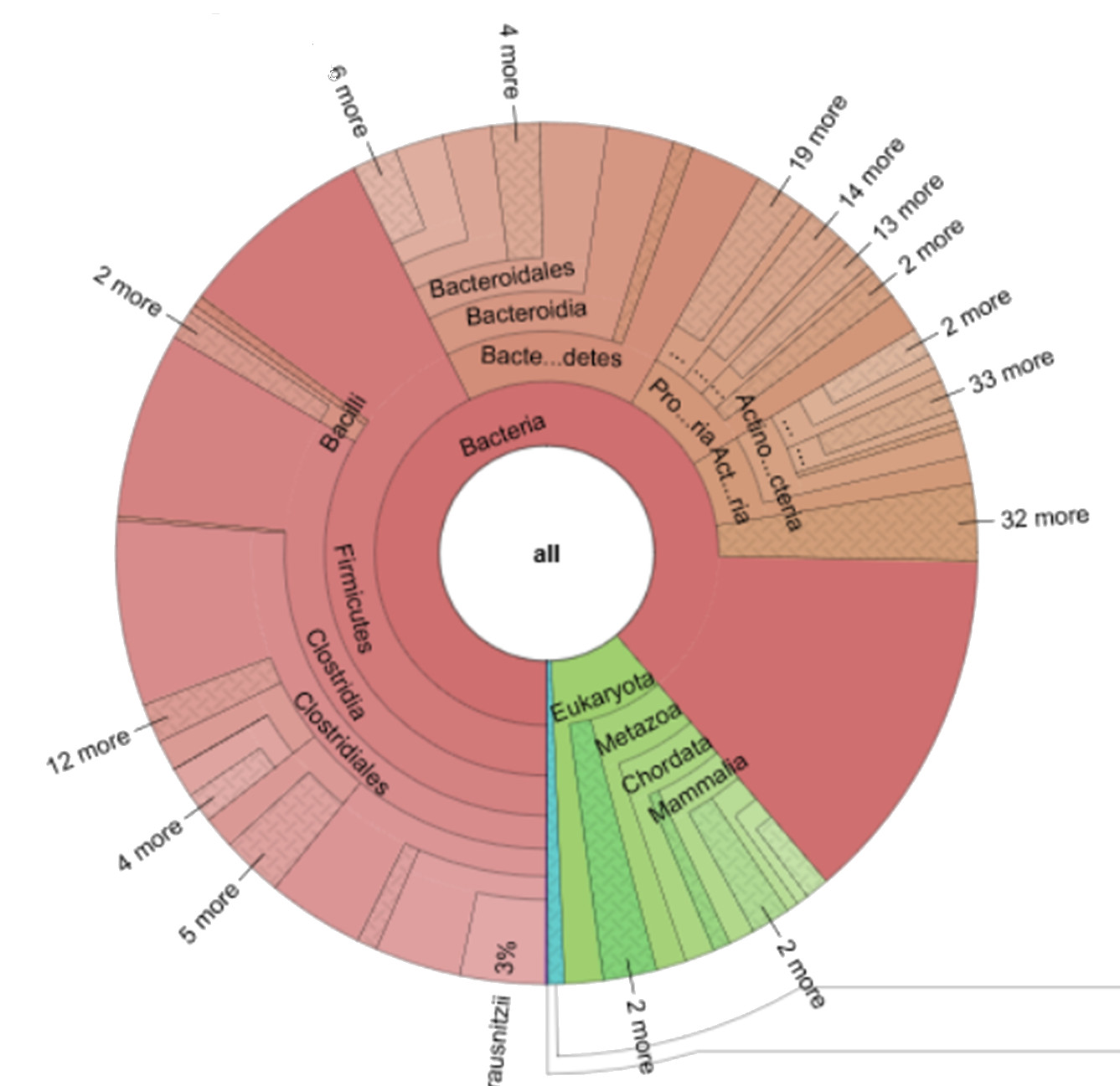
We provide interactive graphical representations of the microbiome and metagenome to allow for a very convenient analysis of the complex data.
Example of a human gut microbiome based on “shotgun” sequencing metagenomics:
(made using the KRONA Software, Copyright)
Metatranscriptomics
To analyze the gene activity of microbiome, we offer metatranscriptomics using adapted sensitive RNA-Seq protocols and bioinformatics.
qPCR
We also offer qPCR services for metagenomics or metatranscripts analysis and design custom primers for the detection of organisms or transcripts using an in-house primer design software pipeline for highest specificity.