Degradome Sequencing by PARE: miRNA target site identification
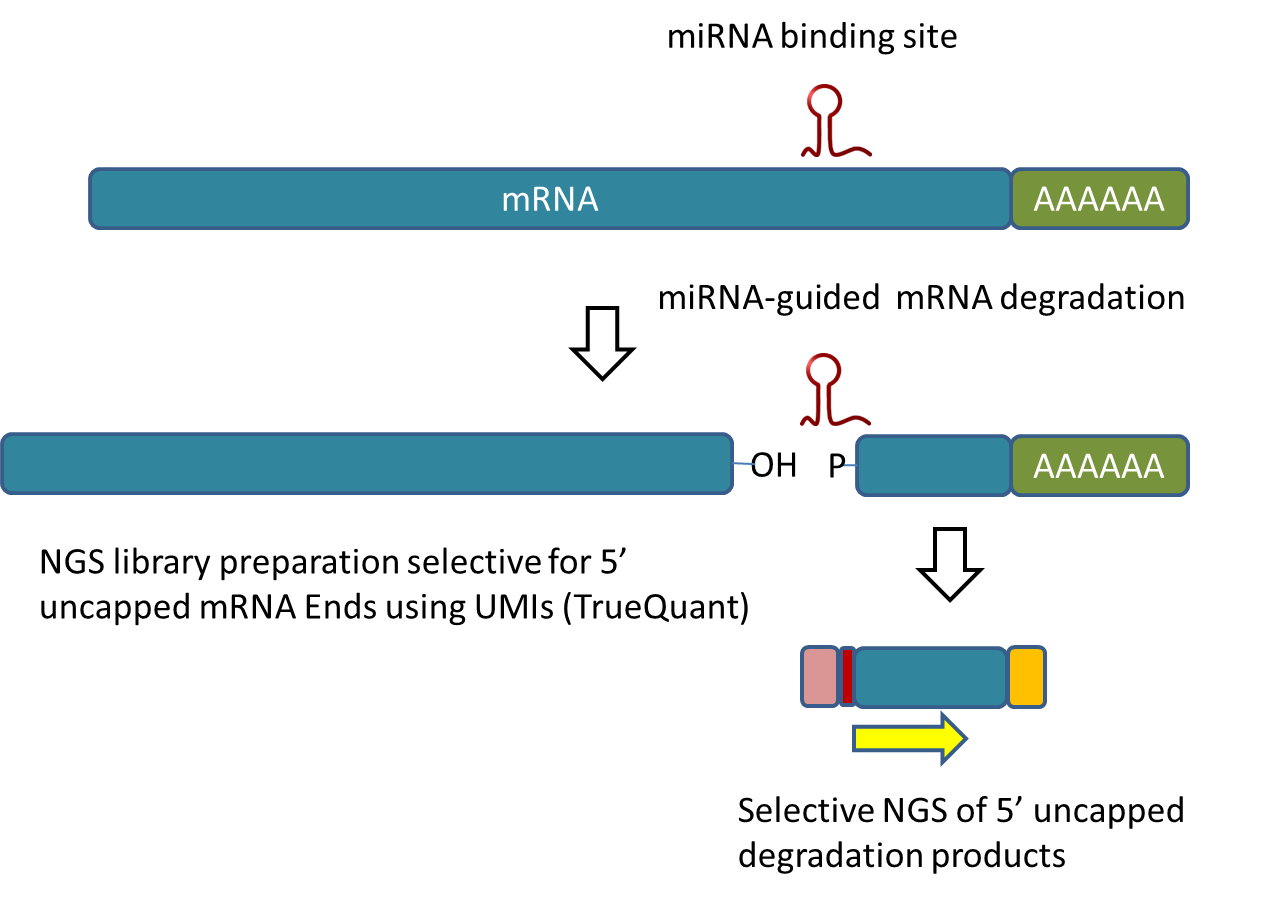
Degradome sequencing (Degradome Seq or PARE = Parallel Analyses of RNA Ends) aims at the identifcation of non-5’capped degradation products of mRNA.
mRNA degradation leads to specific degradation products which carry a phosphate group at the 5′ ends instead of the original 5′ mRNA cap. Our degradome sequencing method is based on selection of phosphorylated and poly-adenylated mRNA pieces which are sequenced from their 5′ ends.
The method is espcially useful for the identification of miRNA-guided degradation products and for miRNA target sites (see picture below).
Since 2008 we use our TrueQuant method for most accurate quantification and PCR bias removal for our degradome seq protocol. Besides the full service (RNA extraction, library generation, sequencing and bioinformatics) we also offer a degradome sequencing kit. Please feel free to contact us for further information at info@genxpro.de.