The MACE-Seq Kit for 3’mRNA Sequencing
Transcriptomics from 0.03 ng of totalRNA
Our highly successful MACE-Seq service is now also available as a kit. It contains all ingredients to perform MACE-Seq. In contrast to other “3’mRNA Seq” or “Tag-Seq” approaches the MACE-Seq Kit includes our patented TrueQuant method (molecular barcodes; Unique Molecular Identifiers, UMIs) to avoid PCR-bias, for the generation of qualitative and quantitative accurate data on any Illumina sequencing machine. MACE-Seq allows for accurate and cost efficient high-resolution gene expression profiling, genotyping in transcripts, allele frequency determination and the analysis of alternative polyadenylation. Up to 386 samples can be processed in parallel.
The low input requirements make it ideal for the analyses of exosomal / EV-cargo or from laser capture microdissected material (LCM). The kit works also very well with low amounts of FFPE-derived RNA.
Bioinformatics
The MACE-seq kit comes with optional bioinformatics-vouchers for a convenient analysis of the data on our servers. As MACE data have some specificities, we have developed an optimized analysis pipeline that you can access using the vouchers. Simply upload your data using the provided voucher-codes to our servers. We provide complete result tables compatible with Excel, as well as plots of your data and Gene Ontology enrichment analyses. The raw and the refined data is accessible via our easily understandable web interface at http://tools.genxpro.net.
Further bioinformatics analyses are possible e.g. SNP-analysis, alternative polyadenylation or allele frequency determination (see Bioinformatics).
Therefore you do not need bioinformatians nor special hardware for the detailed analysis of the complex data.
Analyse between 6 to 384 samples on a MiSeq, HiSeq, NextSeq500 or NovaSeq after 3,5h of sample preparation
The kit is available as 6x MACE version for up to 6 MACE libraries on the Illumina MiSeq platform and a 24-384x version for the Illumina HiSeq or NextSeq.
With further indices, several kits can also be combined for example to reach Y × 24 or Y x 48 samples.
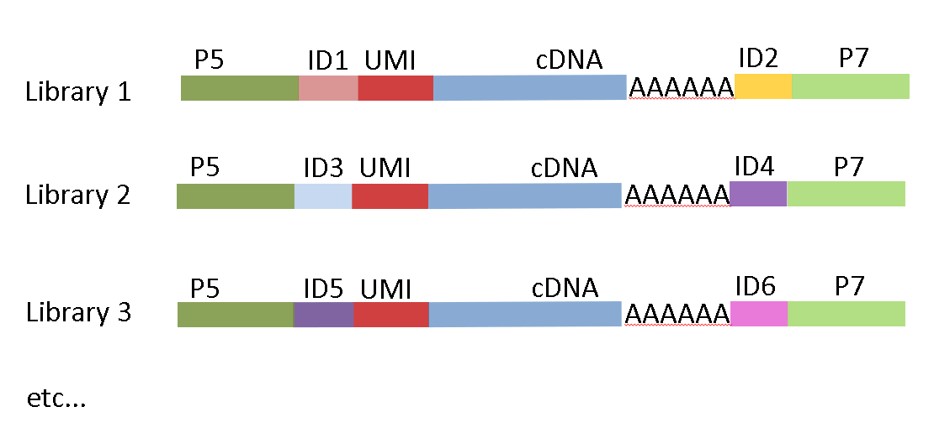
MACE-Seq construct with Unique Dual Indices (UDI)
The latest version of our kit uses unique-dual-indexing to avoid read-misassignment due to “index-hopping” during the illumina sequencing
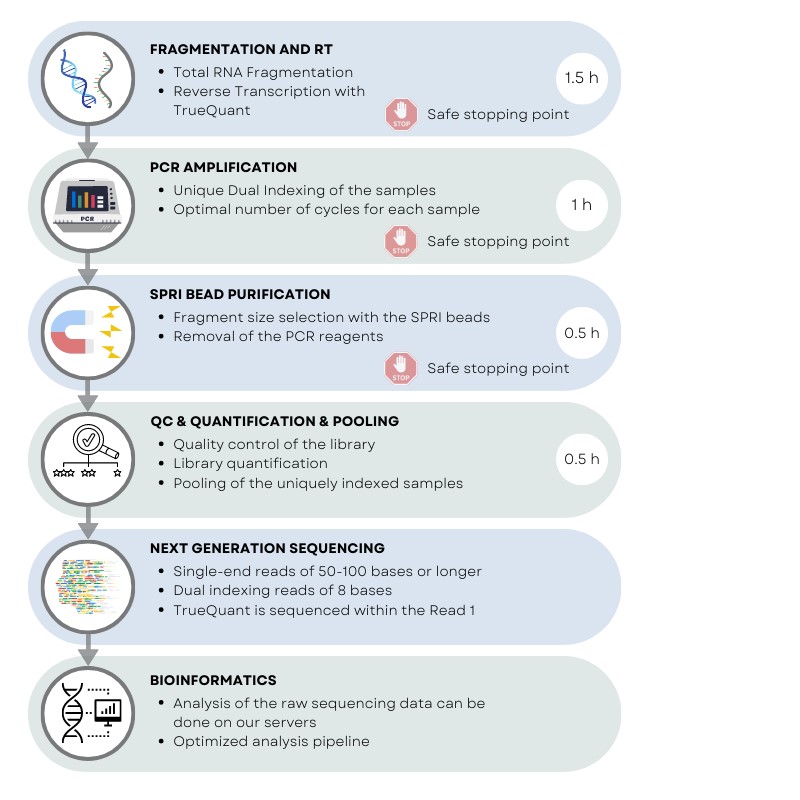
Library preparation workflow for rapid MACE with UDIs. Starting with total RNA, the complete protocol can be carried out in a few hours. The workflow starts with individual fragmentation and reverse transcription of the RNA, followed by indexing-PCR, amplification and SPRI beads purification. After pooling, the indexed libraries can be sequenced on any of Illumina’s NextSeq, NovaSeq, HiSeq, MiSeq, MiniSeq, or iSeq platforms.
Our kit uses UMIs and UDIs in its standard version and optional highly standardized and peer-reviewed published Bioinformatics.
Workflow – Ultra Fast Library Preparation in 3,5 h
Almost no technical variance
the pearson correllation between three technical replicates using MACE-seq from 5 ng of total RNA is shown below. Values of >0,996 proof the very high technical reliability.
| 5 ng – 1 | 5 ng – 2 | 5 ng – 3 | |
| 5 ng – 1 | 1 | ||
| 5 ng – 2 | 0,996987 | 1 | |
| 5 ng – 3 | 0,996446 | 0,99798 | 1 |
Sensitivity and reliability for low input
MACE-seq requires only 0,03 ng of total RNA as input while maintaining consistent transcript quantification. As shown in the table below, the pearson correlation between MACE-seq transcription profiles from 100 ng and 1 ng of total RNA input is 0,96 and 0,87 between data from 100 ng and 0,05 ng total RNA. The lower pearson correlation in the later comparison is due to a reduced number of low-level transcripts in the 0.05 ng sample.
| total RNA | 0.05 ng | 0.1 ng | 0.5 ng | 1 ng | 5 ng | 25 ng | 100 ng |
| 0.05 ng | 1 | ||||||
| 0.1 ng | 0,830518 | 1 | |||||
| 0.5 ng | 0,881877 | 0,923032 | 1 | ||||
| 1 ng | 0,864297 | 0,896227 | 0,939137 | 1 | |||
| 5 ng | 0,864884 | 0,898313 | 0,940447 | 0,995379 | 1 | ||
| 25 ng | 0,871146 | 0,89844 | 0,943703 | 0,978482 | 0,986371 | 1 | |
| 100 ng | 0,867123 | 0,888887 | 0,936261 | 0,959999 | 0,969597 | 0,995612 | 1 |
Sequence your MACE -Seq libraries with other libraries
Because MACE-Sequencing does not require any specific sequencing primers but uses the standard primers for Illumina machines, you can easily sequence your MACE-Seq projects with other projects simultanteously.
We provide unique dual indices for secure identification of the samples.
MACE-Seq for IonTorrent
MACE-Seq is now also available for IonTorrent sequencers. Please ask for further information at info@genxpro.de
Datasheet
Required input material: >0,03 ng totalRNA (can vary depending on tissue and quality of RNA)
Number of samples 1 – 384 (depending on Kit)
Time for sample preparation of 384 samples: 3,5 hours
Bioinformatics optionally included for model- and non-model organisms
Interested? Try it out!
Please send us an e-mail at info@genxpro.de or give us a call at: +49 69 95739710
Customers in Japan: please contact FUNAKOSHI.
NEWS: Our kit received five Stars from “BIOZ“
Service
We also offer MACE-Seq as a service including optional RNA extraction and bioinformatics.
References
Tushev G, Glock C, Heumüller M, Biever A, Jovanovic M, Schuman EM.Alternative 3′ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments.Neuron. 2018 May 2;98(3):495-511
Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science. 2018 May 25;360(6391):881-888.
Agarwal R, Cao Y, Hoffmeier K, Krezdorn N, Jost L, Meisel AR, Jüngling R, Dituri F, Mancarella S, Rotter B, Winter P, Giannelli G.Precision medicine for hepatocelluar carcinoma using molecular pattern diagnostics: results from a preclinical pilot study. Cell Death Dis. 2017 Jun 8;8(6):e2867.
Casado-Díaz A, Anter J, Müller S, Winter P, Quesada-Gómez JM, Dorado G. Transcriptomic Analyses of Adipocyte Differentiation From Human Mesenchymal Stromal-Cells (MSC). J Cell Physiol. 2016 Jun 28
Hofer TP, Zawada AM, Frankenberger M, Skokann K, Satzl AA, Gesierich W,
Schuberth M, Levin J, Danek A, Rotter B, Heine GH, Ziegler-Heitbrock L. slan-defined subsets of CD16-positive monocytes:Characterization of subsets of the CD16-positive monocytes: impact of granulomatous inflammation and M-CSF-receptor mutation. Blood. 2015 Oct 6. pii: blood-2015-06-651331.
Nold-Petry CA, Lo CY, Rudloff I, Elgass KD, Li S, Gantier MP, Lotz-Havla AS, Gersting SW, Cho SX, Lao JC, Ellisdon AM, Rotter B, Azam T, Mangan NE, Rossello FJ, Whisstock JC, Bufler P, Garlanda C, Mantovani A, Dinarello CA, Nold MF. IL-37 requires the receptors IL-18Ra and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal transduction.Nat Immunol. 2015 Mar 2
More references at: https://genxpro.net/publications/
Disclaimer:
The MACE-kit is for research use only.