TrueQuant SmallRNA Seq Kit for Ultra Low Input, GEL FREE, Single Tube Protocol
The GenXPro small RNA sequencing kit (v1.0) can be used to prepare small RNA libraries to be sequenced on Illumina® NGS instruments starting with single-cell amounts of total RNA of 0.01 to 500 ng of total RNA.
The library preparation methodology used in this kit allows an accurate identification and quantification of microRNAs, piRNAs and other small RNAs.
Fast & simple single Tube Protocol, Gel free!
The entire workflow takes place in a single tube. This minimizes losses and increases the efficiency tremendously. Even 0.01 ng of total RNA can be sufficient input material for library preparation.
Less PCR cycles, less duplicates: more output = much lower costs!
Thanks to the highly efficient method to generate a sequencable library from small RNA, usually between 5 and 14 PCR cycles less are needed to obtain the minimum amount needed to load a sequencing lane compared to other standard methods. Our kit therefore often allows to drastically reduce the sequencing costs of smallRNA between 50% and 80% compared to most other methods.
Automation
The single tube protocol strongly facilitates highly paralleled analyses of many samples on liquid handling systems.
TrueQuant Technology
GenXPro small RNA sequencing kit applies the patented TrueQuant Technology that eliminates PCR-derived copies from the generated gene expression data. Following sequencing of the adaptor-ligated and PCR-amplified libraries, only the sequence with highest quality of each TrueQuant adapter plus template-sequence combination are maintained.
Liquid Biopsies from only 200 µl of Plasma
Our protocol allows to generate smallRNA-Seq profiles from down to 200 µl of plasma.
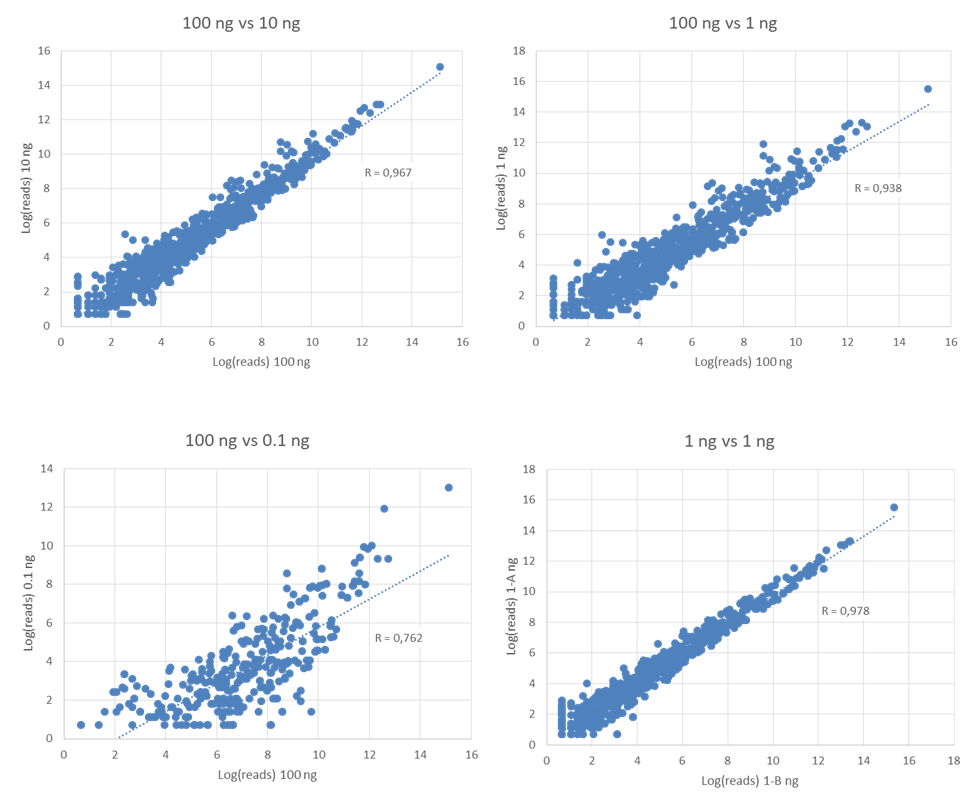
High Technical Reliability and Sensitivity
Scatterplots and pearson correlations (R) of comparisons between different amounts of totalRNA from the same sample.
Protocol for low input amounts, even from strongly degraded total RNA
Because of the single tube protocol that avoids gel purification steps, combined with the TrueQuant method, very low amounts of input material (0.01 ng) even from strongly degraded total RNA is suitable for processing.
CRISPR guide RNA (gRNA) quality control by sequencing
Our TrueQuant smallRNA kit is ideally suited for sequencing for quality control of CRISPR guideRNA (gRNA) and of other RNA constructs including miRNAs or siRNA with a size of up to 100 bps. We also provide powerful solutions for the quality control of longer RNA such as mRNA constructs. For DNA-constructs such as aptamers or plasmids please see https://genxpro.net/aptamers/.
Profit from our well-established nucleic-acid quality control (“NAQC”) analysis pipeline to quantify precisely all variants of the constructs to avoid off-target effects.
We offer the CRISPR guide RNA QC as full service or you can use our TrueQuant smallRNA seq kit and upload the data to our NAQC pipeline for bioinformatics analyses.
Principle and Procedure
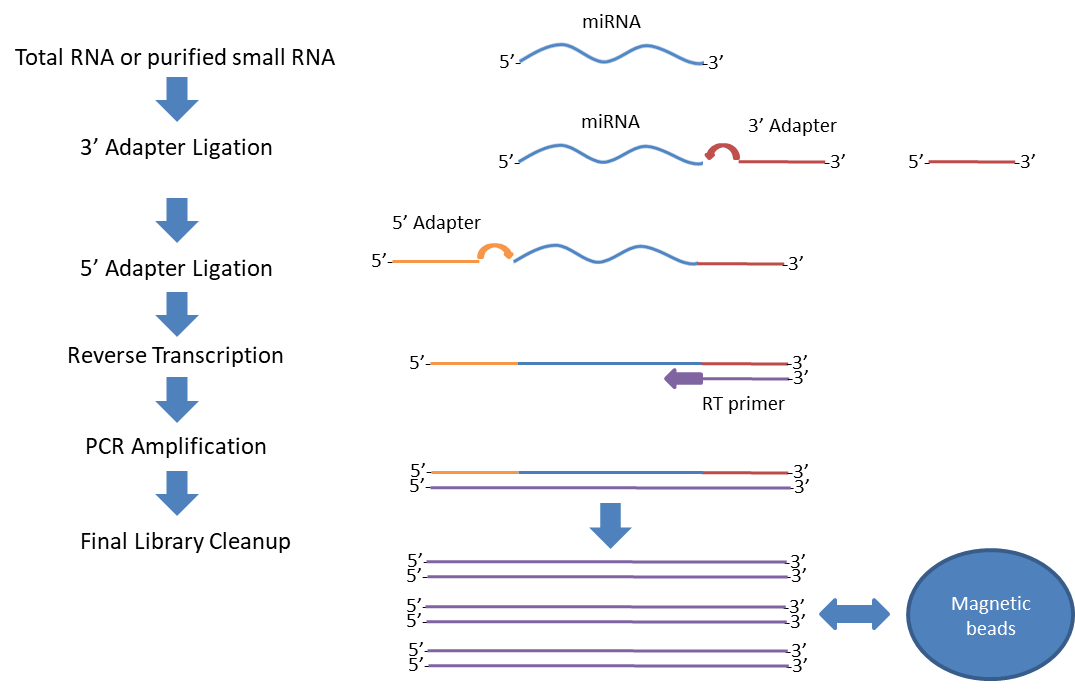 Starting with DNase-treated and quality-controlled total RNA, adapters containing TrueQuant barcodes are ligated to both ends of the smallRNA, reverse transcribed and the final libraries are ready for sequencing on any of the Illumina MiniSeq, MiSeq, HiSeq, and NextSeq platforms. The present kit allows for multiplexing of up to 96 samples.
Starting with DNase-treated and quality-controlled total RNA, adapters containing TrueQuant barcodes are ligated to both ends of the smallRNA, reverse transcribed and the final libraries are ready for sequencing on any of the Illumina MiniSeq, MiSeq, HiSeq, and NextSeq platforms. The present kit allows for multiplexing of up to 96 samples.
Trial possible!
Please send us an e-mail (info@genxpro.de) if you are interested in testing our smallRNA-kit.
Service
We also offer regular and ultra-low input smallRNA seq service . Our service is also available for sequencing of the libraries that were generated with the kit or for bioinformatics analyses of the data.
Price information / Orders / Quotations
Please visit our web portal for a quotation https://tools.genxpro.net/quotation
some References
J Thromb Haemost. 2023;Oct;21(10):2797-2810.
Wecker T, Hoffmeier K, Plötner A, Grüning BA, Horres R, Backofen R, Reinhard T, Schlunck G.MicroRNA Profiling in Aqueous Humor of Individual Human Eyes by Next-Generation Sequencing.Invest Ophthalmol Vis Sci. 2016 Apr 1;57(4):1706-1713.
Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I, Roth WK, Rotter B, Hoffmeier K, Kahl G, Koch I, Theis FJ, Kleeff J, Winter P, Michalski CW. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015 Apr 25;14(1):94.
Disclaimer
The kit is for research use only.


